Introduction to Hyperparameter Tuning in Machine Learning.
Strive for continuous improvement, instead of perfection. - Kim Collins.
Data Scientists and Machine Learning Engineers often contemplate the model and parameters to produce the best accuracy after data preprocessing. Hyperparameter tuning is the process or method used to find out the best parameters for a model. In this article, you will learn the various methods of hyperparameter tuning and their implementation. A working understanding of python and DS libraries like Numpy will be useful in following this article through.
Hyperparameter Tuning using Cross Validation Score Object
Cross-Validation Score is an object in sklearn.model_selection that tunes your model manually by taking in the algorithm, its parameters, the independent features (X), and, the dependent feature (y) of your data. It then performs cross-validation with the given input and returns n number of accuracy scores, where n is the number of splits in cross-validation. See the example below.
Code Example
Import the Pandas and Numpy library to begin with.
import pandas
The BMI index dataset from Kaggle will be used in this article. You can download it here. The goal of this dataset is to predict a person's Body Mass Index(BMI) based on gender, height, and age. In the real world, these features may be insufficient, but for this tutorial, they will suffice.
First, read the dataset and print out the first five rows to see how the data is structured.
bmi_data = pd.read_csv('bmi.csv')
bmi_data.head()
Output
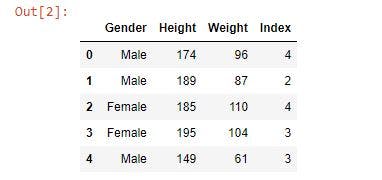
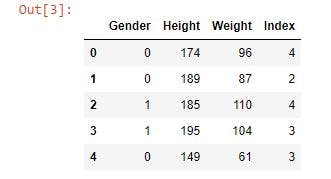
Next, replace the Male and Female data in the gender column with 0 and 1, respectively, as most machine learning algorithms only work on numerical data. Also, display the first five rows using bmi_data.head.
bmi_data = bmi_data.replace(to_replace = ['Male', 'Female'], value = [0,1])
bmi_data.head()
Output
Next, separate the independent and dependent features when training a model. Dependent(y) features are those you want to predict and are dependent on other variables for their values, while your independent(X) features are those used to predict dependent feature(s) values.
Index is the dependent variable in the dataset, while Gender, Height, and, Weight are the independent variables. You can perform this separation using Pandas .iloc[] indexing attribute.
X = bmi_data.iloc[:,0:3] # Independent variable
y = bmi_data.iloc[:,3] #Dependent Variable
Now you are ready to tune your model.
Import the cross_val_score object from sklearn.model_selection.
from sklearn.model_selection import cross_val_score
Next, import the algorithm the Decision Tree Classifier algorithm for training, testing, and building the model.
from sklearn.tree import DecisionTreeClassifier
Next, input the algorithm, its parameters, the independent and dependent features, and the number of splits in cross-validation(cv).
The min_samples_split and max_depth parameters of the Decision Tree Classifier will be varied to find which value(s) give better accuracy.
cross_val = cross_val_score(DecisionTreeClassifier
(min_samples_split = 3,
max_depth = 3),
X, y, cv = 5)
Use Numpy .mean() attribute to display the average of the result.
print(cross_val, np.mean(cross_val))
Output
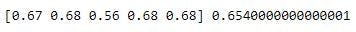
The mean accuracy was 0.654 for a min_samples_split of 3 and max_depth of 3.
Run this same code but with a min_samples_split of 5 and a max_depth of 7, to check if there will be any improvement.
cross_val_ = cross_val_score(DecisionTreeClassifier(min_samples_split = 5, max_depth = 7), X, y, cv = 5)
print(cross_val_, np.mean(cross_val_))
Output
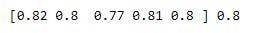
The accuracy went up from an average of 0.654 to an average score of 0.8. Quite impressive.
You could keep on choosing different values for different parameters to see which produces the right fit, but that would take a lot of time. Using a for loop will make things more complex because you would have to write multiple for statements for every additional parameter you choose to add. Luckily, sklearn.model_selection comes with a package called GridSearchCV, which allows you to select as many parameters and values as you want, helping you tune your model with few lines of code.
Hyperparameter Tuning using GridSearchCV
To get started, import GridSearchCV for sklearn.model_selection
from sklearn.model_selection import GridSearchCV
GridSearchCV takes in the model you want to use on your data, followed by a dictionary containing your parameters and their values, then the number of cross-validation splits you want to perform.
Next, create an instance of the GridSearchCV object and input the values stated above to it.
clf_decision_tree = GridSearchCV(DecisionTreeClassifier(), {
'max_depth' : [1,3,5,7,9],
'min_samples_split' : [2,3,4],
}, cv=5, return_train_score=False)
The max_depth parameter takes in 5 values, while the min_samples_split parameter takes in 3. The option provided by GridSearchCV to compute multiple values at a go, makes it more efficient than cross_val_score.
Next, fit the data using the .fit() method and display the result using the .cv_results_ attribute.
clf_decision_tree.fit(X,y)
clf_decision_tree.cv_results_
Output

Pass the result into a dataframe for better visualization using Pandas .DataFrame() method
grid_results = pd.DataFrame(clf_decision_tree.cv_results_)
grid_results
Output
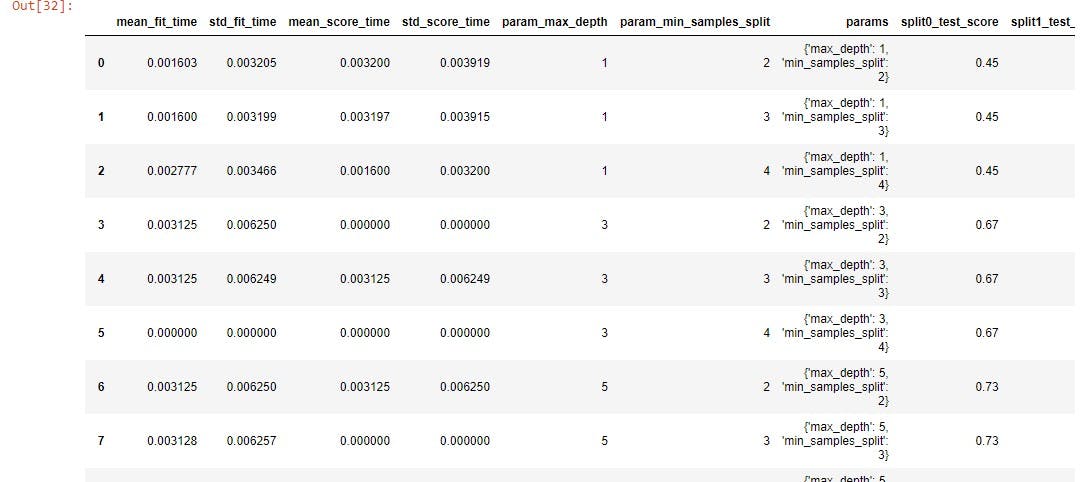
The .cv_results_ attribute return values such as the mean test score, the parameters, individual accuracy of each split performed, and so on. You can view all the columns it returns using the .columns attribute. Not all these values are necessary, just the parameters and their mean test scores. To get this, index the params and mean_test_score features.
grid_results[['params', 'mean_test_score']]
Output
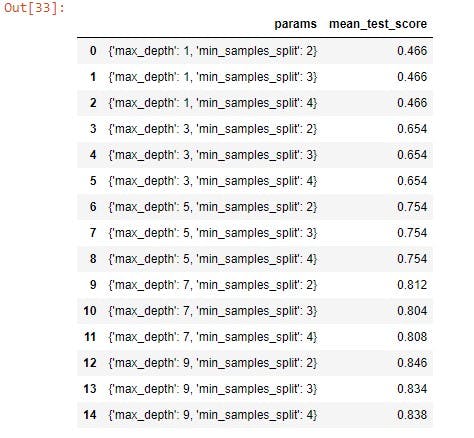
The result above shows the best accuracy is achieved when the max_depth was set to 9 and min_samples_split was set to 2.
GridSearchCV is a great tool to use when there is the confidence that the best accuracy will be gotten using a few sets of values. The limitation of GridSearchCV is that it can't be used for a wide range of values. For example, setting the min_samples_split to take numbers from 1 to 100 will be computationally expensive. To solve this problem, whenever hyperparameter tuning is performed for a wide range of values, RandomizedSearchCV is employed.
Hyperparameter Tuning using RandomizedSearchCV
RandomizedSearchCV works exactly like GridSearchCV except that it tunes the model based on n number of random values where n is the number of iterations you choose.
Get started by importing RandomizedSearchCV from sklearn.model_selection
from sklearn.model_selection import RandomizedSearchCV
Next, create an instance of RandomizedSearchCV and pass in the parameters similar to that of GridSearchCV. The number of iterations parameter n_iter will be set to two. You can use any value of your choice.
random_cv = RandomizedSearchCV(DecisionTreeClassifier(), {
'max_depth' : [1,3,5],
'min_samples_split' : [2,3,4],
}, cv=5,
n_iter = 2
)
Next, fit your data to random_cv and display the results in a DataFrame for easy visualization.
random_cv.fit( X,y)
random_cv_df = pd.DataFrame(random_cv.cv_results_)
random_cv_df
Output
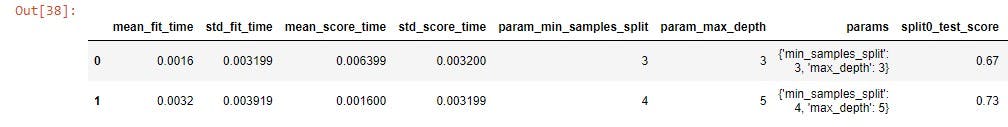
Index the params and mean_test_score values to have a clearer picture of the data.
random_cv_df[['params', 'mean_test_score']]
Output

Using max_depth values of 2 and 9 and `min_samples_split' values of 3 and , give accuracies of 0.58 and 0.838.
Rerun the code.
Output

Repeating again with max_depth values of 3 and 9 and `min_samples_split' values of 4 and 2 to give accuracies of 0.58 and 0.834. The results do not differ so much, but you get the idea.
Choosing the best model with the right Parameters
Finding the appropriate model is as important as finding the best parameters. GridSearchCV will be used to find this best model with the best parameter in this article, though both GridSearchCV and RandomizedSearchCV can be used.
DecisionTreeClassifier, RandomForestClassifier and, AdaBoostClassifier are the three algorithms to be tested on this dataset, along with two of their parameters.
Import the algorithm libraries to get started. You can exclude importing DecisionTreeClassifier as it has been done above.
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier
Next, create a list containing instances of the algorithms and the parameters to be tested embedded in a dictionary.
best_model = [
{
'model' : DecisionTreeClassifier(),
'parameters': {
'min_samples_split' : [1.0, 3, 5],
'max_depth' : [2, 4,6]
}
},
{
'model' : AdaBoostClassifier(),
'parameters': {
'algorithm' : ['SAMME', 'SAMME.R'],
'learning_rate' : [1,2,3]
}
},
{
'model' : RandomForestClassifier(),
'parameters': {
'min_samples_split' : [1.0, 3, 5],
'class_weight' : ['balanced', 'balanced_subsample']
}
}
]
To know more about the parameters of each algorithm.
Next, go through the list and input the model and its parameters to GridSearchCV for tuning of the model. Also, append the best parameters and scores to a list.
best_model_params = []
best_model_scores = []
for model in best_model:
clf_best = GridSearchCV(model['model'], model['parameters'], cv=5, return_train_score=False)
clf_best.fit(X,y)
best_model_params.append(clf_best.best_params_)
best_model_scores.append(clf_best.best_score_)
Next, print the best_model_params and the best_model_scores list
print(best_model_params)
print('')
print(best_model_scores)
Output
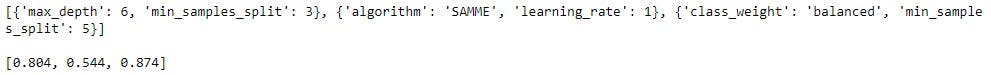
RandomForestClassifier returned the best accuracy with class_weight set to balanced and min_samples_split to 5.
Conclusion
Hyperparameter Tuning is needed when building a model because it gives the optimal parameters that produce the best accuracy. You get an unfair advantage when participating in competitions on platforms like Kaggle and Zindi using Hyperparameter Tuning, placing you higher on the leaderboard.
Moving forward, you can read on other preprocessing methods such as
I hope you learned the basics of Hyperparameter Tuning and how to implement it on data. Tuning your parameters will enable you to get the best out of your model.
If you have any questions, please don't hesitate to reach me on Twitter: @ee_ephraim.

